Discover how to efficiently upload billions of rows into Apache HBase with Apache Spark
HBase is a Big Data database which holds tables of up to billions of rows and millions of columns. It’s client APIs provid lighting fast random read/writes access to its data. It is an open source Apache project which was based on Google’s Bigtable database.
To perform inserts and updates into a table, HBase provides the user-friendly PUT API and the TableOutputFormat class when using a map reduce job. For very large table inserts, the process of bulk loading involves writing a map reduce job to generate the underlying HBase store files (HFiles). These files are directly loaded into the HBase cluster; bypassing the write ahead logs and memstore. This achieves a more efficient and stable process for loading in large volumes of data.
Audience
This tutorial should help professionals working with Big Data systems particularly HBase and Apache Spark, to efficiently and reliably load very large volumes of data on top of an existing table. Readers will also gain an understanding of some HBase architecture and the benefits of using Apache Spark as a map reduce framework.
Prerequisites
Before beginning this tutorial, some knowledge of Hadoop and it’s architecture is required. You will also require access to a running Hadoop cluster if you intend on running this example.
Background
The most common way to generate HFiles is through Hadoop’s native map reduce framework and it’s built in TotalOrderPartitioner class(which we will discuss later on). Although there are existing articles and examples available online, Apache Spark provides a more efficient and eloquent way to perform Big Data map reduce operations.
The following worked example will explain how to generate HFiles using Apache Spark and to bulk load them in a multi region HBase table.
The HBase architecture illustrates that each of the table’s HBase regions has a corresponding store file/HFile on the underlying distributed file system.
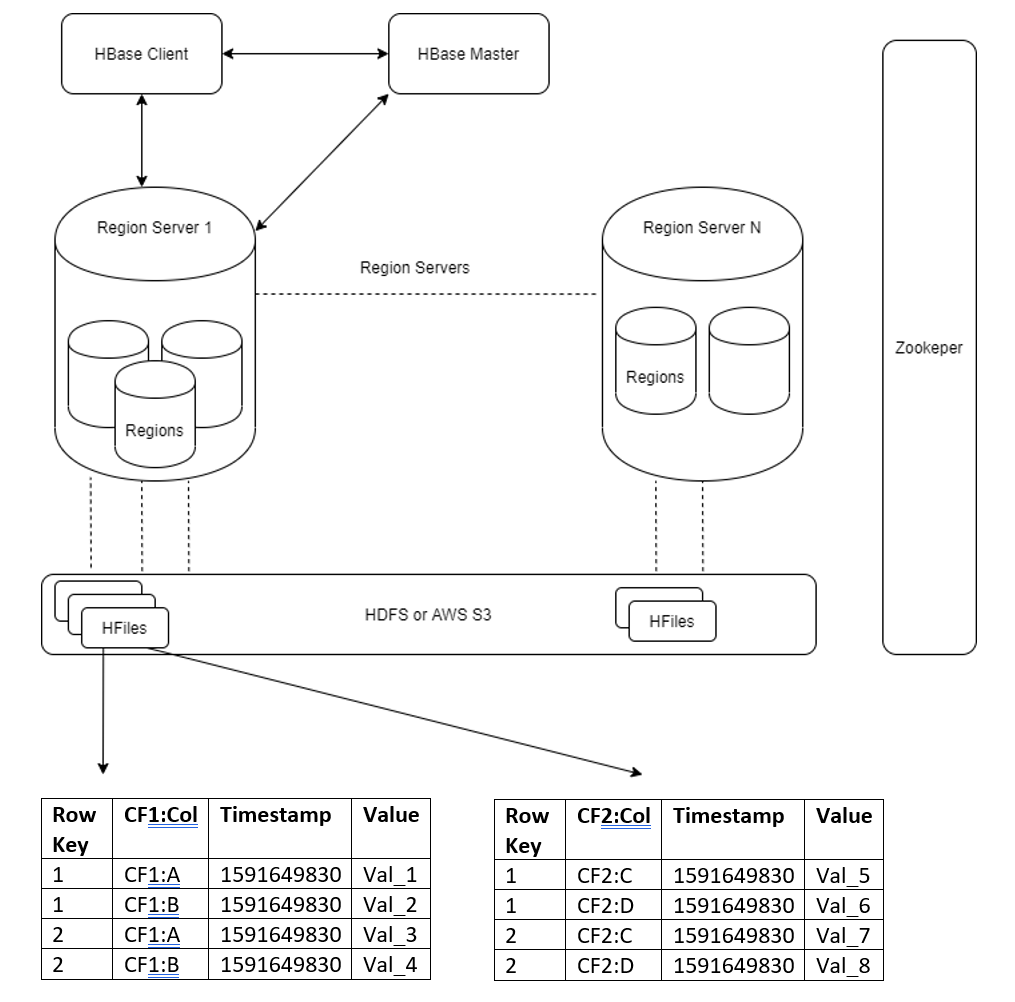
To ensure HFiles are valid and uploaded efficiently:
- The number of HFiles must always match the number of regions for the table.
- HBase sorts its row keys lexicographically from lowest to highest thus each region has a start and end key. Ensure the manually generated HFiles match the start and end row keys for each region.
- Ensure entries in the manually generated HFiles are sorted lexicographically by row key and then column qualifier.
I have created a Spark utility to remove the complexity from the developer who needs to generate HFiles in Apache Spark. Let’s walk through how this utility works.
The first component is essential, it’s the HFile partitioner which is a custom Apache Spark paritioner that partitions the row keys correctly. Rows are assigned to their corresponding region based on start and end keys (rule 2).
The lexicographical sorting of HFile entries and number of HFiles partitions is set in HFileGenerater.scala class.
Our goal is to achieve this table in a multi-region HBase table called ’employees’

Steps
1. Create the empty employees table with a single table split at 4
This will create the table over two regions with entries with row ID <= 4 in the first region and entries with row IDs > 4 in the second region.
hbase(main):015:0> create 'employees', {NAME => 'personal', VERSIONS => 1, COMPRESSION => GZ}, {NAME => 'work', VERSIONS => 1, COMPRESSION => GZ}, SPLITS => ['4']
2. Create employeeDetails.csv file
Input CSV File employeeDetails.csv. This is the small sample data that we want to convert from a csv file to HFiles that can be bulk loaded into the employees table. For my example this is a very small dataset but, in reality, the bulk loading will only be necessary for very large datasets.
id,name,city,title,rating
1,Stephen,Dublin,Accountant,2
2,David,California,Engineer,4
3,Philip,Las Vegas,Accountant,5
4,Glen,Paris,Analyst,4
5,John,Dublin,Security,3
6,Kevin,Dublin,Engineer,4
7,Sophie,London,Architect,53. Create the Spark HFile Custom Partitioner
This partitioner will repartition and sort the our employee data based on the table region splits’ start keys. For more information regarding custom partitioners see the Apache Spark documentation. As previously mentioned, the correct partitioning is fundamental to generating HFiles that can be loaded directly into their corresponding HBase regions. If the partitioning is incorrect, HBase will have to perform very expensive ‘correcting process’ or even fail to generate the HFiles if they are not lexicographical ordered. This does a very similar job to the TotalOrderPartitioner.java which HBase provides from Hadoop map reduce jobs.
import org.apache.hadoop.hbase.io.ImmutableBytesWritable
import org.apache.hadoop.hbase.util.Bytes
import org.apache.spark.Partitioner
import scala.annotation.tailrec
/**
* Partitions and sorts rdd: RDD[(ImmutableBytesWritable, Array[Cell])] so it can be used in the creating of HFiles
* that are intended for bulk loading
*
* Connects to the table regions to read in the start keys for each split and partitions the rdd data accordingly.
*/
class HFilePartitioner(startKeys: Array[Array[Byte]]) extends Partitioner {
override def getPartition(key: Any): Int = {
val rowKey = key.asInstanceOf[ImmutableBytesWritable].get
binarySearch(rowKey, startKeys)
}
override def numPartitions: Int = startKeys.length
override def equals(other: Any): Boolean = other match {
case h: HFilePartitioner => h.numPartitions == numPartitions
case _ => false
}
override def hashCode: Int = numPartitions
/**
* For larger numbers of partitions it's much more efficient to use the binary search with its time complexity of O(log n)
* versus time complexity of O(n) the basic linear search
*/
private def binarySearch(key: Array[Byte], startKeys: Array[Array[Byte]]): Int = binarySearch(key, startKeys, 0, startKeys.length - 1)
@tailrec
private def binarySearch(key: Array[Byte], startKeys: Array[Array[Byte]], fromPosition: Integer, toPosition: Integer): Int = {
val withinRangeValue = (fromPosition: Int) => if (fromPosition < 1) fromPosition else fromPosition - 1
if (fromPosition > toPosition) return withinRangeValue(fromPosition)
val midPointPosition = (fromPosition + toPosition) >>> 1
val comparison = Bytes.compareTo(key, startKeys(midPointPosition))
comparison match {
case 0 => midPointPosition
case c if c > 0 => binarySearch(key, startKeys, midPointPosition + 1, toPosition)
case _ => binarySearch(key, startKeys, fromPosition, midPointPosition - 1)
}
}
}
4. Create a Data Frame from the ‘Employees’ data source
val employeesDF = spark.read
.option("header", "true")
.csv("<path_to_employees_csv>")
5. Convert to RDD
val employeesHFileRdd: RDD[(ImmutableBytesWritable, Array[Cell])] = employeesDF.rdd.map(employee => {
val rowKey = employee.getAs[String]("id")
val name = employee.getAs[String]("name")
val city = employee.getAs[String]("city")
val title = employee.getAs[String]("title")
val rating = employee.getAs[String]("rating")
// 'personal' column families
val nameCell = Cell("personal".getBytes(), "name".getBytes, name.getBytes)
val cityCell = Cell("personal".getBytes(), "city".getBytes, city.getBytes)
// 'work' column families
val titleCell = Cell("work".getBytes(), "title".getBytes, title.getBytes)
val ratingCell = Cell("work".getBytes(), "rating".getBytes, rating.getBytes)
(new ImmutableBytesWritable(rowKey.getBytes), Array(nameCell, cityCell, titleCell, ratingCell))
})
6. Connect to the Employees HBase table
val hBaseConf = new Configuration
hBaseConf.set("hbase.zookeeper.quorum", "<zookeeper_quorem>")
val connection = ConnectionFactory.createConnection(hBaseConf)
val table = connection.getTable(TableName.valueOf("employees")).asInstanceOf[HTable]
7. Instantiate the partitioner
Through using a custom partitioner, we are forced to use Apache Spark’s low level RDD APIs. High level DataFrame APIs don’t provide custom partitioner functionality.
val hFilePartitioner = new HFilePartitioner(table.getRegionLocator.getStartKeys)
8. Create A Utility for Building the HFile RDD using the Partitioner
import org.apache.hadoop.hbase.KeyValue
import org.apache.hadoop.hbase.io.ImmutableBytesWritable
import org.apache.hadoop.hbase.util.Bytes
import org.apache.spark.Partitioner
import org.apache.spark.rdd.RDD
/**
* This is a Utility for converting rdd: RDD[(ImmutableBytesWritable, Array[Cell])] into HFiles
*
* Uses the HFileGenerator utility to convert the delta files data into HFiles using a single shuffle,
* which can be bulk loaded into HBase
*/
object HFileGenerator {
def convertToHFileRdd(rdd: RDD[(ImmutableBytesWritable, Array[Cell])], partitioner: Partitioner) = {
rdd
.repartitionAndSortWithinPartitions(partitioner)
.flatMap(buildHFileEntries)
}
private def buildKeyValueCell(rowKey: ImmutableBytesWritable)(cell: Cell) = {
cell.customTimestamp match {
case Some(ts) => (rowKey, new KeyValue(rowKey.get, cell.columnFamily, cell.columnName, ts, cell.columnValue))
case _ => (rowKey, new KeyValue(rowKey.get, cell.columnFamily, cell.columnName, cell.columnValue))
}
}
private def buildHFileEntries(row: (ImmutableBytesWritable, Array[Cell])) = {
val lexicographicalOrder: (Cell, Cell) => Boolean = (prev, current) => Bytes.compareTo(prev.columnName, current.columnName) < 0
val rowKey = row._1
val cells = row._2
cells
.sortWith(lexicographicalOrder)
.map(buildKeyValueCell(rowKey))
}
case class Cell(columnFamily: Array[Byte], columnName: Array[Byte], columnValue: Array[Byte], customTimestamp: Option[Long] = None)
}
9. Run the Utility
val hFileRdd = HFileGenerator.convertToHFileRdd(employeesHFileRdd, hFilePartitioner)
10. Write the RDD as HFiles in HDFS
val job = Job.getInstance(hBaseConf, "HFile Generator")
// connects to the HBase table and configures the jobs compression, configureBlockSize etc
HFileOutputFormat2.configureIncrementalLoad(job, table, table.asInstanceOf[HTable].getRegionLocator)
hFileRdd.saveAsNewAPIHadoopFile("<hdfs_output_path_for_hfiles>",
classOf[ImmutableBytesWritable],
classOf[KeyValue],
classOf[HFileOutputFormat2],
job.getConfiguration)
11. Inspect the HFiles Structure
We now have two column families and two partitions in each which corresponds to our single table split. If we had more table splits (regions) we would have more partitions in our HFiles.
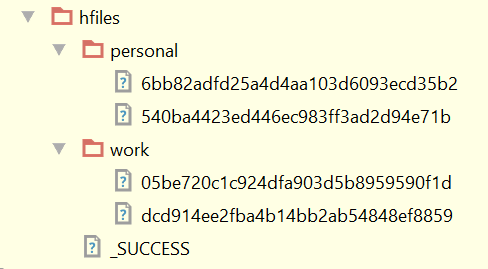
12. Bulk Load HFiles into Employees Table
In the HBase shell bulk load the HFiles into the employee table. I have tested this utility on 10 billion rows and the bulk load time took only seconds.
hbase org.apache.hadoop.hbase.mapreduce.LoadIncrementalHFiles <path_to_hfiles> employees
Final Thoughts
With the strength and popularity of Apache Spark growing each day, more applications will benefit from utilities like this HFile generator. If it would be useful, I will publish it in the Maven repository.
If anyone has any questions or feedback please comment below.
Great post! helped me a lot with Hbase migrations.
LikeLike